Characterization of Carbon Nanotube-Based Probes by Raman Hyperspectral Imaging: Multiplexing and Biodetection
Raman spectroscopy is a powerful technique to identify molecules with their vibrational fingerprints. However, Raman cross-section is usually too low for imaging. In this work by Prof. Richard Martel’s group from the University of Montreal [1], a giant Raman scattering effect is reported in encapsulated and aggregated dye molecules inside single-walled carbon nanotubes (SWNT).
These nanoprobes are based on SWNT and J-aggregated dyes, such as α−sexithiophene (6T), β-carotene (βcar) and phenazine (Ph). Raman probes have the advantages of being stable over long periods of time (weeks and years) and they produce a unique signature with narrow peaks that allows easy multiplexing of 3 probes or more using the same excitation laser energy. This nanomaterial shows a very high Raman scattering cross-section, without any photobleaching or fluorescence background, even at high laser intensities.

In this work, Photon etc.’s hyperspectral Raman imaging platform, RIMA enables the imaging and multiplexing of three different probes with sensitivity down to the single object (Fig 1). The different probes were deposited on a SiOx/Si surface and characterized by taking hyperspectral Raman imaging. Researchers were able to determine the position of each isolated probe (diameters: 1.3 ± 0.2 nm), and identify the co-localized probes (Fig 1 right, Ph and βcar). The sensitivity, efficiency and hyperspectral properties of RIMA were essential to the development of these probes.
The carbon nanotube, which serves as a capsule for the probe, can be covalently functionalized to selectively target biomolecules, such as streptavidin. RIMA’s potential in the detection of probes in a biological context was demonstrated by imaging the βcar probe functionalized with PEG-biotin groups that targeted streptavidin.
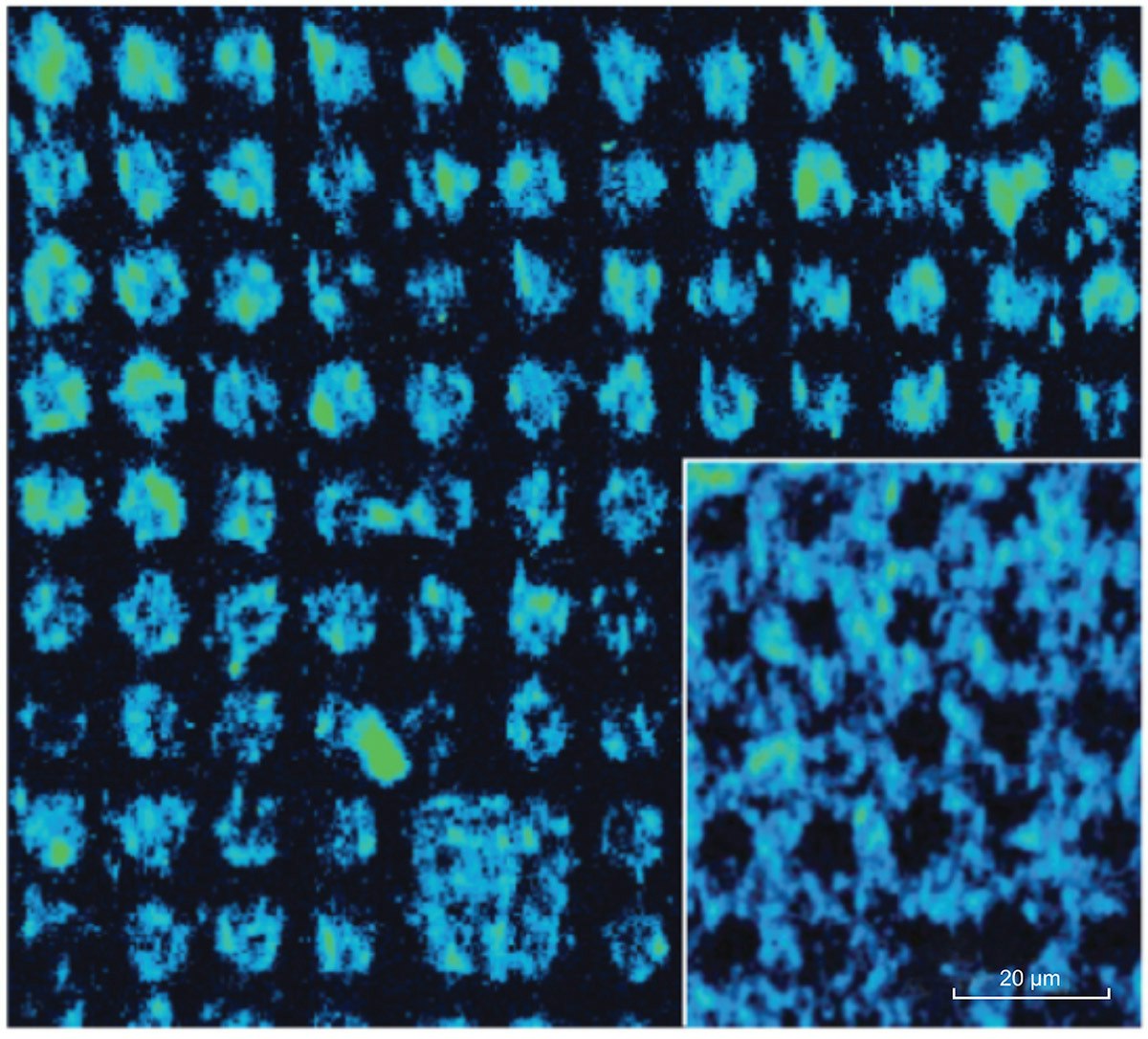
A pattern of 10 μm spots of streptavidin was created by microcontact printing and then incubated with the probes. The pattern was maintained hydrated under a coverslip during imaging and the probes were detected where streptavidin was located. Figure 2 shows Raman hyperspectral images at 1520 cm-1 of two printed surfaces, where streptavidin was deposited either inside (main figure) or around the dots (inset). With a single acquisition, a sample area of 133 x 133 μm2 was studied using RIMA with laser excitation at 532 nm. Damages to the samples were also limited due to a uniform illumination over the portion of the sample in the field of view. In terms of spectral resolution and large surface area imaged, RIMA provided hyperspectral images in a much shorter time than conventional point-by-point mapping Raman imagers.
Raman hyperspectral imaging is a powerful technique to study a wide range of materials, from nanopatterned surfaces to biological systems. Because of its high throughput, RIMA allows the acquisition of spectrally resolved maps of large-area samples, without damaging the surface.
For more information, contact info@photonetc.com
[1] E. Gaufrès et al., Giant Raman scattering from J-aggregated dyes inside carbon nanotubes for multispectral imaging, Nature Photonics, 2014, 8 72-78. Copyright 2014 Macmillan Publishers Ltd.